Lawlor Lab
Unraveling the mysteries of Ewing sarcoma
The goal of the Lawlor Lab is to discover how hijacking of normal developmental biologic programs contributes to the origin and progression of childhood cancer. Our research is focused on Ewing sarcoma, an aggressive bone and soft tissue tumor for which new cures are needed. Through investigations of tumor biology, we are identifying new therapeutic opportunities. Current areas of interest are tumor heterogeneity, epigenetic and metabolic reprogramming, and discovering how these processes contribute to tumor metastasis.
Current Research and Projects
 Tumor heterogeneity
Tumor heterogeneity
Sarcomas are complex tissues that are comprised of both cancer cells and normal cells, as well as extracellular matrix. Crosstalk and collaboration among all components of the tumor is necessary for local tumor growth as well as metastasis to distant sites. Importantly, our research has shown that Ewing sarcoma tumor cells are very dynamic and they change phenotype rapidly in response to signals from their microenvironment. It is clear from our studies, as well as others, that these changes in cancer cell phenotype contribute to the tumor heterogeneity that drives disease relapse, metastasis, and emergence of drug resistant disease. Using diverse tumor model systems and state-of-the-art genomic, epigenomic, and functional assays, we are elucidating the molecular mechanisms that cause tumor cells to change states.
Our most recent work has focused on tumor microenvironment crosstalk and how Wnt/beta-catenin and TGF-beta signaling in discrete tumor-cell subpopulations collaborate to alter the local extracellular matrix, promoting tumor angiogenesis and disease progression.
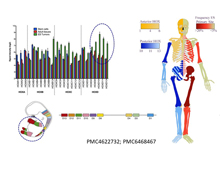
Enhancer reprogramming of posterior HOXD genes is an epigenetic hallmark of Ewing sarcoma. We have shown that hijacking of HOXD13 regulation specifically coopts developmental transcription programs that are essential for tumorigenicity.
Epigenetics
Ewing sarcomas are characterized by the presence of gene fusions, most commonly EWS-FLI1, often without evidence of any other gene mutations. As such, changes in cell phenotype that promote metastatic progression are largely driven by non-genetic mechanisms. We are studying how disruption of normal epigenetic regulation of developmental programs leads to tumor initiation and tumor progression. In particular, we are focused on how epigenetic hijacking of developmental transcription factors such as HOX genes leads to malignant transformation of normal stem cells. In addition, we are studying how these processes work in established sarcoma cells to switch chromatin states allowing cells to adapt to new environments and to stress. Understanding the fundamental biology of epigenetic processes in Ewing sarcoma will lead to new treatment strategies that employ less toxic, tumor-targeted drugs (i.e. inhibitors of bromodomain proteins, HDACs, menin, and other epigenetic modifying agents).
Metabolic reprogramming
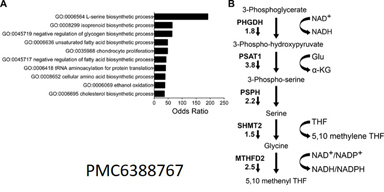
Inhibition of the scaffolding protein menin in Ewing sarcoma leads to metabolic rewiring, with specific loss of serine synthesis pathway (SSP) gene expression and activity. This creates a metabolic vulnerability that we hypothesize can be therapeutically exploited.
The process of rapid proliferation and tumor expansion exposes tumor cells to multiple sources of internal and external stress. Tumor cells have adapted ways to maintain cellular homeostasis that allow them to survive and thrive under conditions that would destroy less resilient cells. Among the most important of these adaptation mechanisms is metabolic reprogramming. By rewiring their metabolic pathways, cancer cells are able to maintain redox and amino acid homeostasis and epigenetic stability, while generating the extra energy and biomaterials that are necessary to sustain their growth. Our current studies of Ewing sarcoma are investigating how serine biosynthesis and amino acid flux contribute to tumor progression. Importantly, these studies have identified critical connections between the epigenome and metabolic reprogramming that we hypothesize are necessary for metastasis.

Elizabeth Lawlor, MD, PhD
Dr. Elizabeth Lawlor completed her MD at McMaster University and her clinical training in pediatric hematology-oncology and bone marrow transplantation at BC Children’s Hospital in Vancouver, Canada. Her passion to discover new cures for childhood cancer led her back to school, where she completed her PhD in pathology and laboratory medicine at the University of British Columbia. She then moved to the University of California, San Francisco to finish her post-doctoral training and then began her career as an independent investigator in 2004 at Children’s Hospital Los Angeles and the University of Southern California. In 2010, Lawlor moved to the University of Michigan, where her lab continued to focus its research on basic and translational studies of pediatric solid tumors, in particular Ewing sarcoma. While at Michigan, she also dedicated her energies to a second passion – graduate and post-graduate education – serving as director of the Cancer Biology PhD graduate and T32 training programs and as associate director of education and training at the Rogel Cancer Center.
Lawlor joined Seattle Children’s and the Ben Towne Center for Childhood Cancer and Blood Disorders Research in June 2020 and currently serves as associate director for the center. She is also a professor of Pediatrics at the University of Washington and adjunct professor in the Department of Laboratory Medicine and Pathology. The overall goal of her research is to discover and define differences between normal developmental and sarcoma biology that will enable discovery of targeted therapeutics that are less toxic to growing children.
Lawlor’s lab has been continuously funded by grants from the National Institutes of Health/National Cancer Institute, American Association for Cancer Research/Stand Up to Cancer, Alex’s Lemonade Stand Foundation, St. Baldrick’s Foundation, the V Foundation, as well as numerous smaller funding agencies. As a dual-trained MD and PhD physician-scientist, she is committed to educating the next generation of cancer researchers and to creating a diverse pipeline of physicians and scientists who will lead us to a better tomorrow for children who are diagnosed with cancer.