Bacteriophage
Bacteriophages (phages) have been minimally used in emergency clinical applications for skin and soft tissue infections and disseminated bacterial infections, but not to-date as an aerosol strategy against pulmonary mycobacteria. One limitation in the field preventing these advancements is the availability of a reproducible preclinical model of aerosol phage delivery that can be leveraged for efficacy testing. Importantly, our preliminary proof-of-concept results1 (read the paper) suggest that prophylactic aerosol delivery of phage can significantly reduce bacterial burden after challenge with M.tb. This work is done extensively in partnership with the Hatfull Lab at the University of Pittsburg. Support for the mycobacteriophage program has come from NIAID grant funding (now complete) and by the Bill and Melinda Gates Foundation (active).
Our bacteriophage interests include:
- Using high throughput screening methods to evaluate the lytic capacity of phage candidates against pathogenic bacteria, including M.tb, NTM and bacteria known to affect persons living with Cystic Fibrosis. Our systems allows testing in a range of strains and conditions, including hypoxia, titrations of MOI, in combination with antibacterial drugs and over a range of timepoints. We believe evaluating therapeutic phage candidates across a physiologic conditions will help facilitate translation to the clinic.
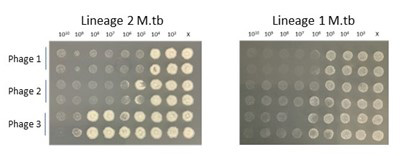
- We are dedicated to developing reproducible preclinical aerosol models of delivery and answer longstanding biological questions about phage treatment in vivo. This includes basic host immunological outcomes of repeated phage treatment, route of delivery and pharmacodynamics/kinetics. Our data suggest that repeated aerosol delivery limits the development of anti-phage humoral immunity and deposits higher titers in the airway when compared to intravenous treatment (read the paper).
These preclinical models are currently being leveraged to evaluate a cocktail of phages designed to treat M.tb in partnership with TASK in South Africa under the Phages4TB consortium, supported by BMGF.
-
The Pacific Northwest region has contributed a minor fraction of mycobacteriophages to the Actinobacteriophage Database at PhagesDB.org (a Hatfull Lab managed resource). Our group performs regular phage hunts using environmental samples collected throughout the year to try and fill this void.

Samples containing prospective phage are screened against nonpathogenic and subsequently pathogenic bacterial isolates. Hits are sequenced and deposited with Phagesdb.org. We subscribe to the model that science should be equitable and accessible so we regularly extend phage hunt invitations to university student groups, scientific and non-scientific staff at CGIDR as well as visiting scientists and interns.
Questions or interest in the bacteriophage program should be directed to Sasha Larsen Akins, Ph.D. ([email protected]).
