Explore Our Research
Proteomic and Genetic Insight Into Axon Termination, Synapse Maintenance and Autophagy
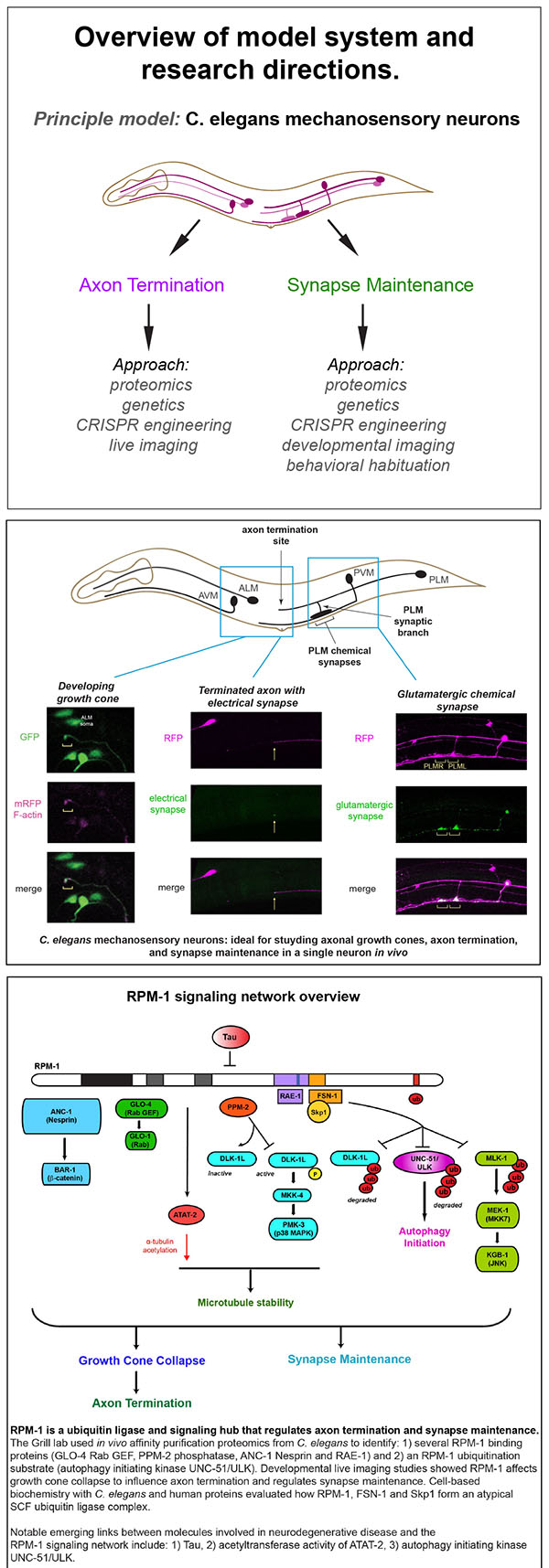
The Grill Lab has made important contributions to understanding signaling networks that regulate axon termination, synapse maintenance and behavioral plasticity. We rely upon C. elegans mechanosensory neurons as our principal in vivo model system. Importantly, much of our progress stems from my group’s status as a leader in C. elegans neural proteomics, which we often use as a starting point for discovery science. Recently, affinity purification proteomics laid the foundation for our exciting discovery that ubiquitin ligase activity inhibits autophagy in the nervous system.
We are leading efforts to understand a conserved signaling network required for both axon termination and synapse maintenance, two fundamental events in nervous system development and stability. At the heart of this network is Regulator of Presynaptic Morphology 1 (RPM-1), a ubiquitin ligase and signaling hub.
Our proteomics work established RPM-1 as the first example of a ubiquitin ligase that acts as a signaling hub in the nervous system. Live imaging and developmental time course approaches have allowed us to study the cellular processes that RPM-1 regulates. Behavioral outcomes indicate that perturbing the RPM-1 signaling network affects behavioral plasticity to repeated touch due to effects on glutamatergic chemical synapses.
In recent work, we used C. elegans proteomics and a ubiquitin ligase biochemical “trap” to identify RPM-1 ubiquitination substrates. This unbiased approach identified the autophagy initiating kinase UNC-51/ULK as an RPM-1 substrate. Genetic, transgenic and CRISPR editing/engineering approaches demonstrated that RPM-1 ubiquitin ligase activity restrains initiation of autophagy in mechanosensory neurons and broadly in the nervous system of C. elegans.
This is just the beginning of what will be a leap forward in neural proteomics in the coming years!
Using Forward Genetics to Identify Regulators of Opioid Sensitivity and Tolerance
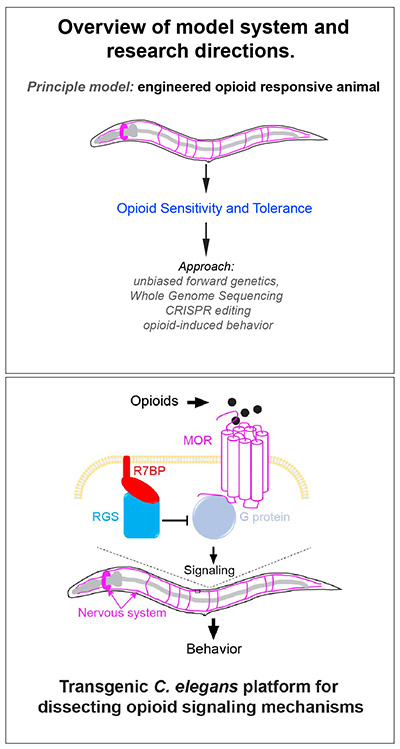
We have spent the past decade pioneering an innovative new line of investigation that aims to understand how u-opioid receptor (MOR) signaling, sensitivity and tolerance are regulated. The implications of this work are far reaching, as MOR is the principle GPCR that mediates analgesia and addiction to opioids.
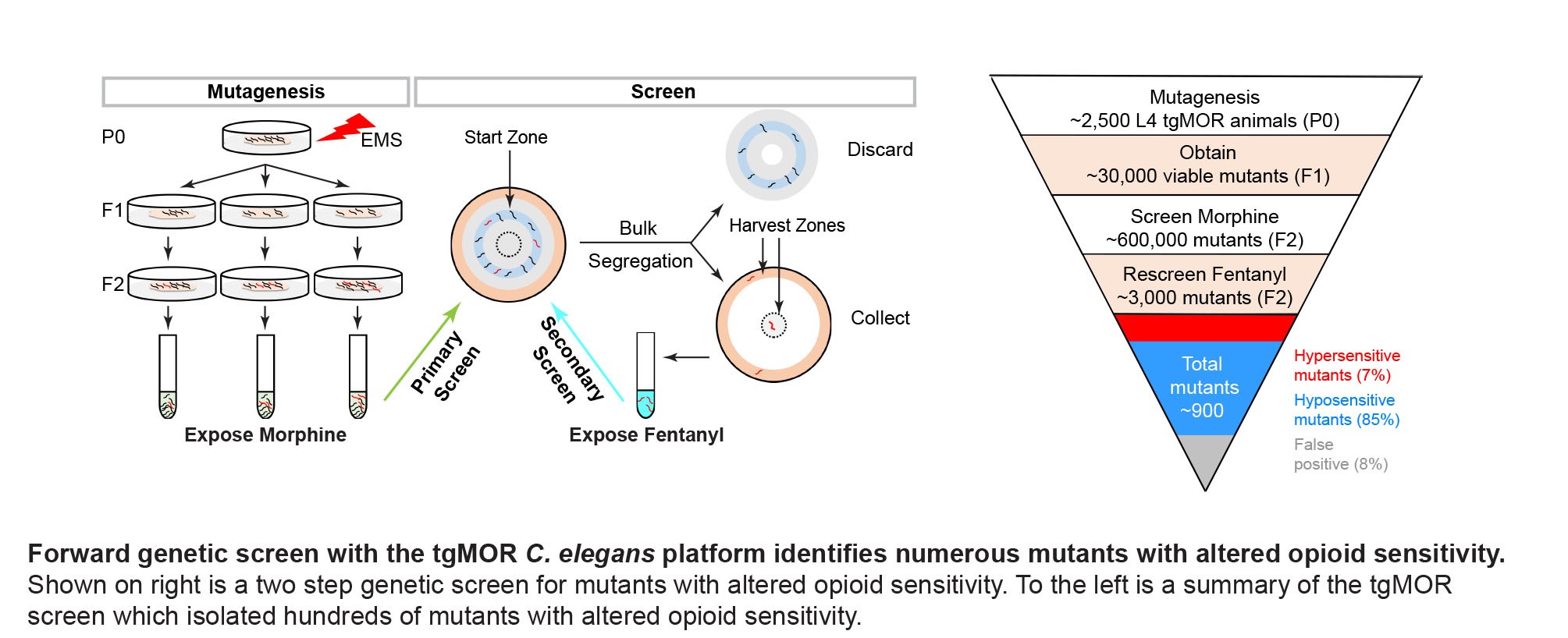
We engineered the first transgenic C. elegans model of MOR signaling (tgMOR), which has robust behavioral responses to opioids. Combining the tgMOR platform with unbiased, forward genetics allowed us to identify numerous, novel regulators of MOR signaling and trafficking that affect opioid sensitivity and tolerance. One example is the orphan GPCR FRPR-13 we identified in C. elegans and its mammalian ortholog GPR139. FRPR-13 and GPR139 functions as a conserved anti-opioid system from C. elegans through mammals. This is the first time that forward genetics has been used to study opioid sensitivity and tolerance in any system. Our unpublished work indicates there is much dark biology behind MOR signaling and potentially a larger network of unknown signaling territory that MOR orchestrates to mediate responses to opioid drugs.
We are also now poised to embark upon developing powerful new engineered behavioral models of GPCR signaling using our C. elegans platform.
Our work is made possible in part by a generous gift in honor of Timothy Jackson.
Regulation of Inhibitory GABA Neuron Function
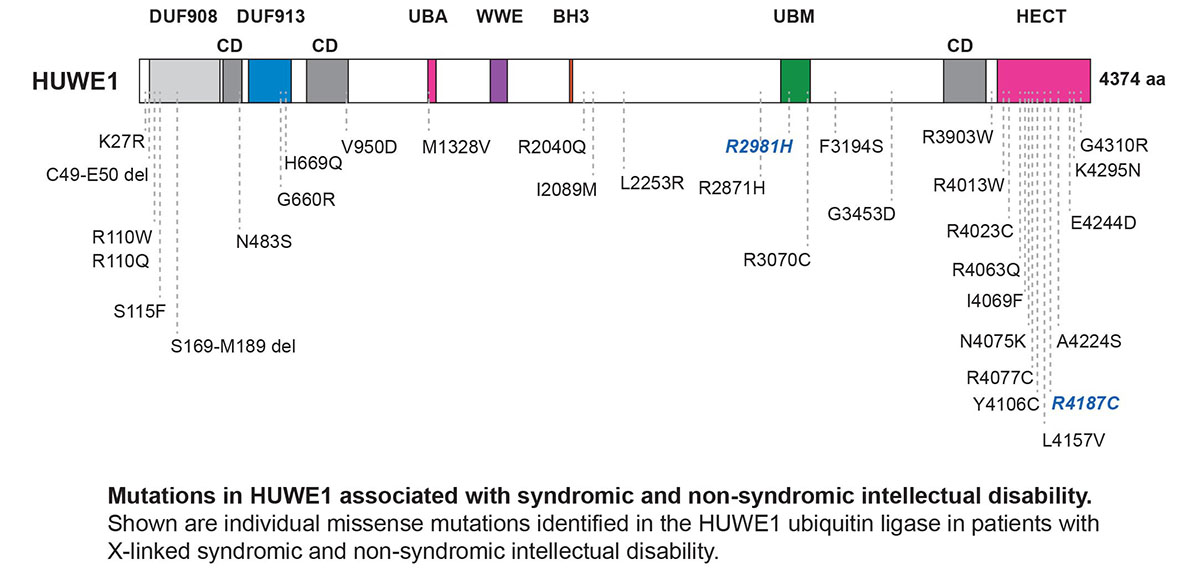
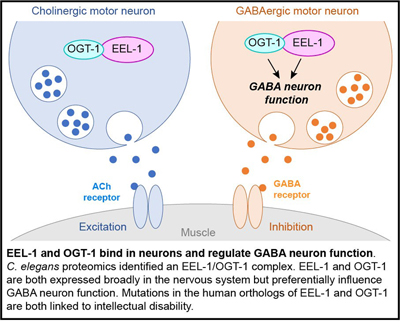 Another ubiquitin ligase that the Grill Lab studies is called EEL-1 in C. elegans and HUWE1 in mammals. Our interest in HUWE1 is driven on two levels. 1) Better understanding how ubiquitin ligase activity shapes inhibitory GABA neuron function. 2) Strong, growing evidence that human genetic changes that reduce or increase HUWE1 function result in syndromic and non-syndromic forms of intellectual disability.
Another ubiquitin ligase that the Grill Lab studies is called EEL-1 in C. elegans and HUWE1 in mammals. Our interest in HUWE1 is driven on two levels. 1) Better understanding how ubiquitin ligase activity shapes inhibitory GABA neuron function. 2) Strong, growing evidence that human genetic changes that reduce or increase HUWE1 function result in syndromic and non-syndromic forms of intellectual disability.
Our work has demonstrated that EEL-1 regulates presynaptic GABAergic transmission, and affects E/I balance in a simple model circuit, the C. elegans motor circuit. To determine how EEL-1 regulates presynaptic GABAergic transmission, we have deployed affinity purification proteomics using C. elegans to identify EEL-1 binding proteins. The initial discovery from these efforts was the finding that EEL-1 forms a complex with the O-GlcNAc transferase OGT-1. Our results using automated behavioral tracking of C. elegans locomotion in liquid, and pharmacological manipulation of the worm motor circuit showed that OGT-1, like EEL-1, affects GABA neuron function. Our work provided the first evidence that EEL-1 and the EEL-1/OGT-1 complex affect GABA neuron function. The biomedical implications of our discoveries are heightened by evidence that genetic changes in human HUWE1 result in intellectual disability, and emerging links between genetic changes in OGT and intellectual disability.
We are now moving forward with both proteomic and genetic screens to further understand how HUWE1 shapes inhibitory GABA neuron function.