Projects
Genetic Risk for Type 1 Diabetes
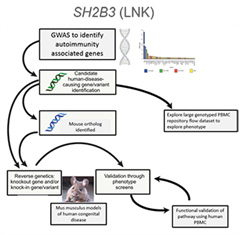
Type 1 diabetes (T1D) is a polygenic disease caused by T-cell mediated destruction of pancreatic β-cells, resulting in life-long insulin dependence. Historically, the majority of T1D patients have been diagnosed at stage 3 of disease development, after significant β-cell destruction, hyperglycemia, and the presence of other clinical symptoms. Genetic risk variants have been defined in an effort to identify high-risk individuals or predict T1D progression, allowing for earlier intervention to preserve β-cell mass. A common genetic variant (present in at least 5% of a population) can get enriched get enriched to help fight a specific microbe (infection) or adjust to a new change in the environment. This same variant may also have unintended side effects like autoimmune disease.
Allenspach Lab researchers combined a genetic association study comparing subjects with type 1 diabetes and controls with similar genetic backgrounds and looked through all common variants across our population to find those enriched for type 1 diabetes. The Allenspach Lab discovered a strongly associated variant (rs3184504) located in the gene SH2B3, encoding the adaptor protein SH2B3 (also termed LNK). This adaptor dampens down cytokine signaling and is highly expressed in blood cells. The genetic region was enriched (selected) in Northern European populations and nearly absent in African and Asian populations that all carry the “ancestral allele” or the allele not associated with diabetes, similar to non-human primates. Through studying primary blood samples from humans and developing a novel small-animal model to study this risk during stress (infection) and in models of autoimmunity, the project goal is to better understand how this critical adaptor protein shapes the immune response in the presence of this precise genetic variant.
- Funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- Read the study findings in the journal Diabetes.
Understanding the Underlying Variants Leading to Autoimmunity or Predisposition to Infection
 The Allenspach Lab’s patient-directed research aims to confirm and understand how particular genetic mutations can lead to disease. Rare genetic conditions are often characterized by early onset and severe symptoms, yet can be challenging to diagnose given limited knowledge or the uncharacterized nature of the disease. Researchers in the Allenspach Lab apply next-generation sequencing paired with deep immunophenotyping approaches to aid in unsolved cases. By studying rare or early onset cases, Allenspach Lab researchers might accelerate diagnoses and/or acquire knowledge more broadly about the immune system.
The Allenspach Lab’s patient-directed research aims to confirm and understand how particular genetic mutations can lead to disease. Rare genetic conditions are often characterized by early onset and severe symptoms, yet can be challenging to diagnose given limited knowledge or the uncharacterized nature of the disease. Researchers in the Allenspach Lab apply next-generation sequencing paired with deep immunophenotyping approaches to aid in unsolved cases. By studying rare or early onset cases, Allenspach Lab researchers might accelerate diagnoses and/or acquire knowledge more broadly about the immune system.
Active projects:
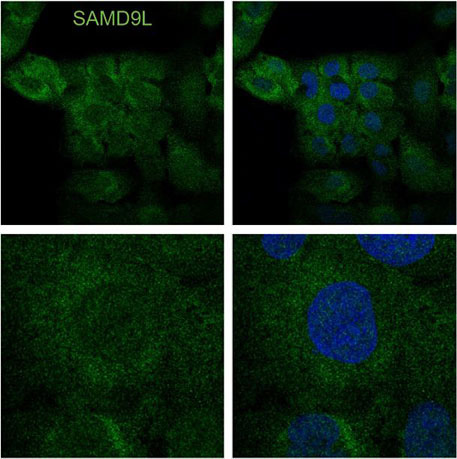
- The Allenspach Laboratory identified a gain-of-function or enhanced human variant that blocks protein synthesis in human cells with surprisingly cell-type specific effects. SAMD9L is an interferon-induced tumor suppressor implicated in a spectrum of multi-system disorders including risk for myeloid malignancies and immune deficiency. This project has focused on understanding how blocks in translation lead to unique stress responses, cell survival pathways and the role of SAMD9L as an antiviral protein.
- Funded by the Brotman Baty Institute, Ataxia-Pancytopenia Foundation and SCRI.
- Funded by the Brotman Baty Institute, Ataxia-Pancytopenia Foundation and SCRI.
- Ask new questions of known immune system disorders such as deeply studying a family with numerous siblings with RAG2-deficiency. RAG2 is an essential gene that when missing causes Severe Combined Immunodeficiency (SCID) or “boy-in-the-bubble” disease. However, less severe (hypomorphic) variants can lead to immune dysregulation and even autoimmune disease. In deeply studying blood samples from several affected siblings longitudinally over 30 years, the Allenspach Lab employed repertoire analysis of T cells and B cells to gain insight into the immune system with this perturbation.
Allenspach Lab researchers demonstrated a range of pediatric and older adult presentations and highlighted the importance of genetic validation studies for an accurate diagnosis. The same genetic changes in three affected siblings in a family prompted more detailed analysis, including a combined approach requiring multiple functional assays to define pathogenicity of one of the family’s genetic variants. This study will help geneticists and immunologists avoid pitfalls in the analysis of rare genetic variants during initial diagnosis.
Memory B cells, ones previously involved in an immune response, can appear in the peripheral blood and look a lot like a circulating cells from the spleen — marginal zone B cells — responsible for making most of the baseline IgM antibodies in humans. This lack of reliable markers limits study of genetic variants. In one of the patients described in this study, these were dramatically expanded, enabling single-cell RNA sequencing to explore these cells further and compare to those pooled from healthy individuals.
- How do mosaic and somatic variants impact the immune system? Mosaic genetic changes are only carried in a portion of cells in the body occurring after the single cell (post-zygotic) stage. Somatic variants are thought to be acquired after birth. Having only a portion of your cells with the pathogenic variant can cause unique phenotypes. The Allenspach Lab team is exploring the immune phenotype in patients with ALPS, or autoimmune lymphoproliferative syndrome (FAS gene), RALD, or Ras-associated lymphoproliferative disease (NRAS/KRAS genes) and reversion mutations in IL2RG gene. The detection of these changes is particularly challenging and can significantly alter the symptoms in each patient.
