Maves Lab
 The Maves lab investigates skeletal muscle and heart development, with the goal of making discoveries that lead to new treatments for muscular dystrophy and heart disorders. We use the zebrafish as an animal model because of advantages for genetic manipulations, in vivo imaging and drug screening.
The Maves lab investigates skeletal muscle and heart development, with the goal of making discoveries that lead to new treatments for muscular dystrophy and heart disorders. We use the zebrafish as an animal model because of advantages for genetic manipulations, in vivo imaging and drug screening.
Projects
We currently have three main projects:
- We are using a zebrafish model of Duchenne muscular dystrophy (the dmd mutant strain) to investigate the developmental biology and epigenetics of DMD as well as to identify new drug therapies for DMD. We also work closely with Dr. David Mack’s lab (https://iscrm.uw.edu/faculty/david-l-mack/) to take advantage of human iPSC and rat models of DMD.
- We are investigating the mechanisms of skeletal muscle fiber-type differentiation. This work will eventually help us understand why certain types of muscle fibers are more susceptible to muscular dystrophy.
- We are using CRISPR genome screening and precision editing in zebrafish to engineer mutations in genes that have been implicated in human congenital heart defects. Recently our screening has identified several new genes required for heart development. Our goal is to gain insight into the complex genetics of congenital heart defects.
Using a 3-model approach to study the developmental biology and epigenetics of DMD and to identify novel drug therapies
Duchenne muscular dystrophy (DMD) is a muscle wasting disease caused by mutations in the DMD gene, which encodes dystrophin. There are presently no cures. The current standard of care is corticosteroid treatment, which delays the progression of muscle dysfunction but also has serious side effects. Although DMD is a progressive disease, there is growing evidence for early-stage defects in myogenesis and gene expression in DMD. By understanding how these early developmental defects initiate and how they drive DMD pathology, we may be better positioned to identify and utilize DMD therapies. The Maves Lab is using a zebrafish DMD model to investigate the developmental biology and epigenetics of DMD as well as to identify new drug therapies for DMD. We recently identified novel epigenetic small molecules that improve muscle degeneration in DMD zebrafish (https://doi.org/10.1186/s13395-020-00251-4). We also work closely with Dr. David Mack’s lab (https://iscrm.uw.edu/faculty/david-l-mack/) and their human iPSC and rat models of DMD. The long-term goal of this project is to take advantage of our three DMD models to identify conserved (and, thus, likely important) molecular and developmental defects underlying DMD. We also expect that potential therapeutic approaches that are beneficial to all three models will have more likelihood of success in DMD clinical trials.
Understanding skeletal muscle fiber-type differentiation

One focus of our lab is understanding the mechanisms behind how skeletal muscle cells differentiate into a specific muscle fiber type, either fast twitch or slow twitch. We are using zebrafish and mouse models to study the functions of factors such as Pbx homeodomain transcription factors and how they control muscle fiber-type development.
Certain muscular dystrophies preferentially affect either fast or slow twitch muscle fibers. In human and mouse models of Duchenne Muscular Dystrophy (DMD), fast muscle fibers are more susceptible to damage than slow fibers. It is not clear how fiber-type identity confers susceptibility or resistance to muscular dystrophy. A goal of this project is to test a fundamental hypothesis: that factors that promote slow muscle differentiation will ameliorate the effects of DMD. Recent studies, including work from our lab, now create an opportunity to directly test whether fiber-type modulation is a viable therapeutic approach for muscular dystrophies. We take advantage of zebrafish models to address whether factors that regulate fiber-type differentiation enhance or suppress the zebrafish dmd muscle degeneration phenotype. Our approach is to manipulate fiber-type regulators that function early in development in dmd zebrafish embryos. We are also working to identify new epigenetic factors that regulate muscle fiber type. We are screening epigenetic chemicals for their abilities to enhance or suppress the zebrafish dmd phenotype. We expect that this project will identify genetic and epigenetic regulators of muscle fiber-type identities that confer susceptibility or resistance to muscular dystrophy. This project is aimed at an ultimate goal of manipulating skeletal muscle fiber type as a treatment for muscular dystrophy.
Using zebrafish and CRISPR to understand the mechanisms of heart development and the genetics of congenital heart defects
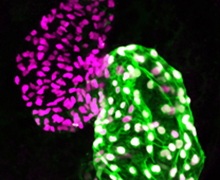 Our long-term goal with this project is to achieve a precise understanding of the genetic and transcriptional mechanisms underlying early heart development. Our group has shown that Pbx and Meis homeodomain transcription factors are critical regulators of heart development in zebrafish. However, the mechanisms underlying Pbx/Meis function in heart development and congenital heart defects are not understood. We are using CRISPR genome editing to engineer mutations in zebrafish pbx and meis genes to determine their functions in heart development. We are particularly interested in understanding how Pbx and Meis factors regulate the differentiation of cardiomyocytes. Because the transcriptional regulation of cardiomyocyte differentiation is highly conserved, our zebrafish work will have relevance for human heart development and disease. We are one of the first labs to use CRISPR to demonstrate a function for a human SNP in zebrafish, a PBX gene SNP associated with congenital heart defects (https://doi.org/10.1242/dmm.035972). Our studies aim to improve our understanding of how transcriptional regulatory factors like Pbx and Meis contribute to heart development and congenital heart defects.
Our long-term goal with this project is to achieve a precise understanding of the genetic and transcriptional mechanisms underlying early heart development. Our group has shown that Pbx and Meis homeodomain transcription factors are critical regulators of heart development in zebrafish. However, the mechanisms underlying Pbx/Meis function in heart development and congenital heart defects are not understood. We are using CRISPR genome editing to engineer mutations in zebrafish pbx and meis genes to determine their functions in heart development. We are particularly interested in understanding how Pbx and Meis factors regulate the differentiation of cardiomyocytes. Because the transcriptional regulation of cardiomyocyte differentiation is highly conserved, our zebrafish work will have relevance for human heart development and disease. We are one of the first labs to use CRISPR to demonstrate a function for a human SNP in zebrafish, a PBX gene SNP associated with congenital heart defects (https://doi.org/10.1242/dmm.035972). Our studies aim to improve our understanding of how transcriptional regulatory factors like Pbx and Meis contribute to heart development and congenital heart defects.
In addition, we are using CRISPR genome screening in zebrafish to test the functions of a new set of genes that have been implicated in human congenital heart defects. Recently our screening has identified several new genes required for zebrafish heart development. We are using protein-protein interaction networks to form hypotheses about how these new genes work with other known heart development genes. Our goal is to gain insight into the complex genetics of congenital heart defects.
Selected Publications
A novel chemical-combination screen in zebrafish identifies epigenetic small molecule candidates for the treatment of Duchenne muscular dystrophy.
Farr GH 3rd, Morris M, Gomez A, Pham T, Kilroy E, Parker EU, Said S, Henry C, Maves L. Skelet Muscle. 2020 Oct 15;10(1):29.
PMID: 33059738
Functional testing of a human PBX3 variant in zebrafish reveals a potential modifier role in congenital heart defects.
Farr GH 3rd, Imani K, Pouv D, Maves L. Dis Model Mech. 2018 Oct 18;11(10):dmm035972.
PMID: 30355621
BMP and FGF signaling interact to pattern mesoderm by controlling basic helix-loop-helix transcription factor activity.
Row RH, Pegg A, Kinney BA, Farr GH 3rd, Maves L, Lowell S, Wilson V, Martin BL. Elife. 2018 Jun 7;7:e31018.
PMID: 29877796
Lipid Nanoparticle Packaging Is an Effective and Nontoxic mRNA Delivery Platform in Embryonic Zebrafish.
Patton C, Farr GH 3rd, An D, Martini PGV, Maves L. Zebrafish. 2018 Jun;15(3):217-227.
PMID: 29369745
"Muscling" Throughout Life: Integrating Studies of Muscle Development, Homeostasis, and Disease in Zebrafish.
Goody MF, Carter EV, Kilroy EA, Maves L, Henry CA.
Curr Top Dev Biol. 2017;124:197-234.
PMID:28335860
Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease.
Talbot J, Maves L.
Wiley Interdiscip Rev Dev Biol. 2016 Jul;5(4):518-34. Review.
PMID:27199166
Pbx4 is Required for the Temporal Onset of Zebrafish Myocardial Differentiation.
Kao RM, Rurik JG, Farr GH 3rd, Dong XR, Majesky MW, Maves L.
J Dev Biol. 2015;3(4):93-111.
PMID:26770887
Conversion of MyoD to a neurogenic factor: binding site specificity determines lineage.
Fong AP, Yao Z, Zhong JW, Johnson NM, Farr GH 3rd, Maves L, Tapscott SJ.
Cell Rep. 2015 Mar 31;10(12):1937-46.
PMID:25801030
The HDAC Inhibitor TSA Ameliorates a Zebrafish Model of Duchenne Muscular Dystrophy.
Johnson NM, Farr GH 3rd, Maves L.
PLoS Curr. 2013 Sep 17;5.
PMID:24459606
A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development.
Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE.
Cell. 2012 Sep 28;151(1):221-32.
PMID:22981225
Faces of Research
Dr. Maves discusses the lab’s work and some of the goals of our research in this video:
Dr. Maves talks about her work and her interest in science in this video:
Partnership Opportunity
Principal Investigator

Lisa Maves, PhD
Dr. Lisa Maves is a principal investigator at Seattle Childrens Research Institute and an associate professor of Pediatrics at the University of Washington School of Medicine. She received her PhD from the University of Washington and completed postdoctoral research at the University of Oregon and at Fred Hutchinson Cancer Center. She was a staff scientist at Fred Hutchinson before joining Seattle Children's Research Institute in 2012.
