Obado Lab
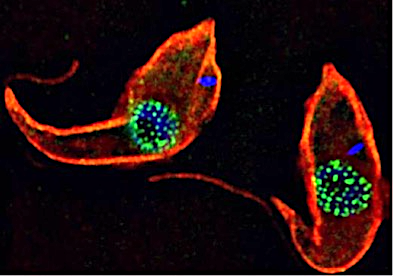
The focus of the Obado Lab, led by Sam Obado, PhD, is understanding the functional relationship between the control of gene regulation, RNA export and the nuclear pore complex (NPC), which is the sole mediator of nucleocytoplasmic transport. Among our model systems are trypanosomes, a diverse family of protozoans that are obligatory parasites of invertebrates, vertebrates and plants. Trypanosomes (including Leishmania) cause major public health and economic problems in the developing world and are a growing problem in the southern states of the U.S. in the form of Chagas disease. Trypanosomes are also excavates, early diverging eukaryotes that exhibit non-canonical biological traits, a number of which differentiate them from their vertebrate hosts. Examples include extensive RNA editing, polycistronic transcription and trans-splicing. Thus, studying their cell biology gives deep insights into which systems are conserved, and which are lineage-specific adaptations, across the Eukarya.
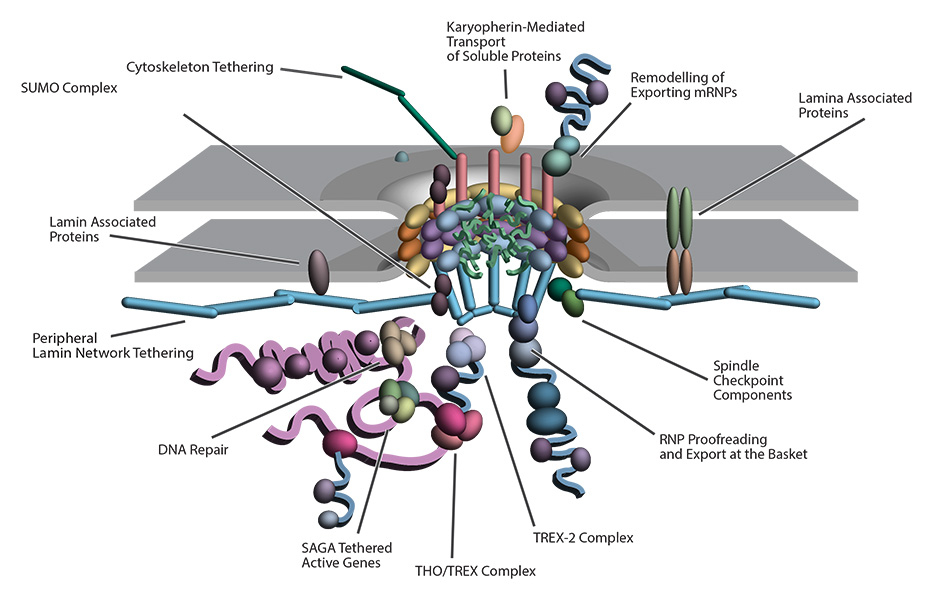
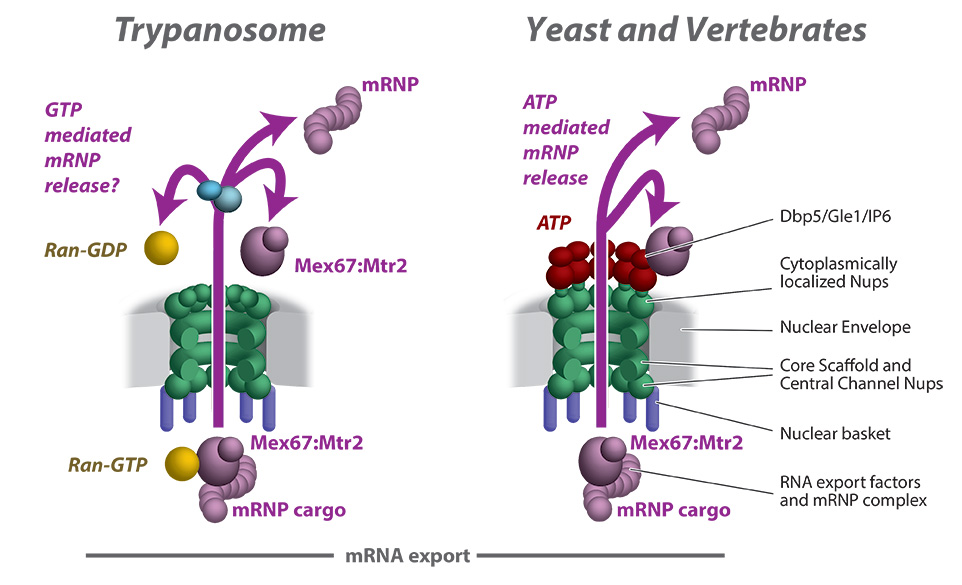
The central dogma of molecular biology highlights the flow of genetic information; that DNA is transcribed into RNA in the nucleoplasm and exported through the nuclear pore complex (NPC), to the cytoplasm, where it is translated into protein. However, several components of the transcription-export complex in trypanosomes are cryptic, and the machinery that drives mRNA export in trypanosomes is evolutionarily divergent from those of its vertebrate hosts. Furthermore, the architecture and composition of the trypanosome NPC contains several atypical features, the most unique of which are related to RNA export. Thus, despite being ubiquitous to all eukaryotes, trypanosomes help show us that the NPC has several species-specific subunit arrangements tuned to the biology of each organism. The divergent trypanosome NPC architecture raises questions about the universality of RNA export in eukaryotes and our understanding of RNA export control, which has largely been studied in opisthokonts, such as yeast and vertebrates. This emphasizes Dobzhansky's idea that biology makes sense only in the light of evolution, with trypanosome research shedding light on conserved cell biology principles.
Work in Dr. Obado’s laboratory focuses on uncovering key components of the mRNA export pathway and determining the structure function relationships between nucleocytoplasmic transport and the divergent trypanosome NPC. The goal of the lab is to utilize protein-protein interactome mapping with cell and structural biology to unravel and analyze the composition of the transcription-export complexes and uncover new RNA export pathways in these parasites of public health importance.
Interested in joining the Obado Lab?
Please send your CV to Samson.Obado@seattlechildrens.org.